Research
Last Update 10/8/14
1. Analyzing protein interactions by luminescence
in living plant cells
 |
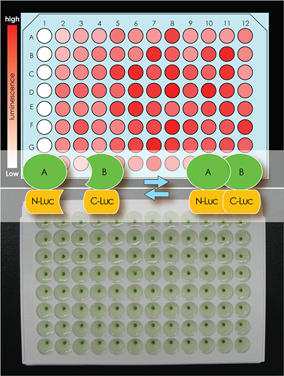
We developed
the split luciferase complementation assay to study
protein-protein interactions in plant protoplasts in 96-well
plates. In this assay, the N- and C-terminal fragments of Renilla
reniformis luciferase are translationally fused to a
protein pair of interest, respectively. When the protein pair
interacts with each other, split luciferase emits luminescence
that can be measured by a microplate luminometer. DNA vectors
expressing the protein pair are constructed through in vitro DNA
recombinant reactions. This system is useful for investigators
who are interested in studying dynamics of protein-protein
interactions in living plant cells. More details can be found in
our publications.
|
| The split luciferase complementation assay |
2. Fatty acid traffic in green microalgae
 |
Plants and green microalgae synthesize triacylglycerols (TAGs) using fatty acids synthesized in plastids and acyl-CoA pooled in endoplasmic reticulum (ER). TAG synthesis and fatty acid traffic in green microalgal species recently generated considerable interest because TAGs have a high potential as renewable fuels. The long-term goal of this project is to develop microalgae that efficiently metabolize fatty acids through modification of the traffic control within the cells. We found Chlamydomonas reinhardtii, a model green microalga, forms novel compartments that accumulate exogenously added fatty acids in the cytoplasm, designated fatty acid-induced microbodies (FAIMs). We are analyzing the biological function of the FAIMs. Some of the results from this project can be found in our publications. |
| Chlamydomonas algae forming FAIMs |
3. Modeling the vesicle traffic in plant cells
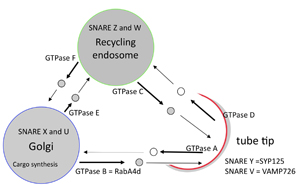 |
The long-term goal of this project is to develop methods to manipulate vesicle traffic in cells of interest by taking the systems biology approach. Arabidopsis pollen expresses only 30 % of genes encoded in the genome and the expressed genes are enriched in sequences that are associated with vesicle trafficking machinery. Thus, Arabidopsis pollen tube growth is a good model to study the molecular mechanism of vesicle trafficking at single cellular levels. As a first step, a mathematical model that describes the flow of molecules in Arabidopsis pollen tubes was constructed. Some of the results from this project can be found in our publications. |
| A diagram of vesicle trafficking translated into ordinary differential equations |
4. Dynamics of organelle interactions in plant
cells
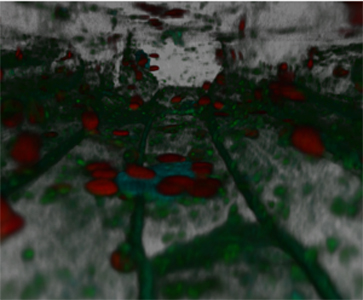 |
Nuclei, plastids, and mitochondria move dynamically to maintain proper cellular function. The interactions among these organelles can be identified and analyzed by tracking their movements simultaneously. To study the dynamics of these organelle-organelle interactions in plant cells, transgenic Arabidopsis, designated Kaleidocell, was engineered. Kaleidocell plant cells contain nuclei, plastids, mitochondria, and plasma membranes that are genetically tagged with cyan, red, yellow, and green fluorescent proteins, respectively. Optically sectioned images of Kaleidocell root epidermis are obtained at a submicron resolution to reveal in living cells the interaction between nuclei, plastids and mitochondria. A three-dimensional model is generated to understand localization of these organelles in the cells. Furthermore, time-lapse observations of three-dimensional projections of the root epidermal cells are generated to track movements of these organelles. The seeds of the Kaleidocell line are available from ABRC. Some of the results from this project can be found in our publications. |
| Inside view of Kaleidocell |